Jeffrey M. Dick
Geochemistry · Thermodynamics · Genome evolution · Microbiomes · Cancer
I am a multidisciplinary researcher investigating fundamental connections from geochemistry and genomes to microbiomes and cancer. To integrate data and theory, I have developed tools for chemical and thermodynamic analysis of omics data. This new view of evolution as a chemical process promotes the application of geochemical concepts to important problems in geobiology and human health.
My data visualization portfolio is now available.

Thermodynamic data and applications
First released more than ten years ago (Geochemical Transactions, 2008), the free CHNOSZ R package for thermodynamic calculations and diagrams for geochemistry has seen continued growth and maintenance (Frontiers in Earth Science, 2019). Recent updates include functions for making diagrams with multiple metals (Applied Computing and Geosciences, 2021).
The OBIGT database in CHNOSZ has thermodynamic data from many published papers. My own contributions include estimates of thermodynamic properties relevant to aromatic maturity parameters in organic geochemistry (Geochimica et Cosmochimica Acta, 2013) and group contribution parameters for proteins that account for temperature and pH effects (Biogeosciences, 2006). When deployed at the genome scale, thermodynamic calculations indicate that overall protein synthesis from inorganic precursors is exergonic (releases energy) in the mixing zone of seawater and vent fluid at hydrothermal systems hosted in ultramafic rocks (Journal of Geophysical Research: Biogeosciences, 2021 ).
From metagenomes to multi-omics
How are microbial communities shaped by gradients of temperature, redox, and water availability?
Proteins in various functional classes and phyla exhibit parallel trends of carbon oxidation state in a hot-spring metagenome (PLOS One, 2011 and 2013). But it's not just about redox; two chemical metrics (ZC and nH2O) of proteins vary along environmental redox gradients and salinity gradients (Frontiers in Microbiology, 2019; Biogeosciences, 2020).
Oxygen and water, bugs and us.
 The healthy human microbiome is shaped by the chemical characteristics of different body sites.
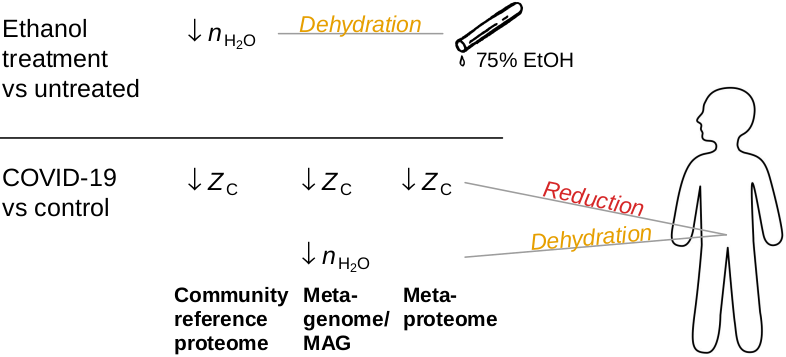
Dysbiosis in COVID-19 is associated with more reduced microbial proteins, informing about how the body's internal chemical environment changes in disease
(preprint: Dick, 2023).
The healthy human microbiome is shaped by the chemical characteristics of different body sites.
Dysbiosis in COVID-19 is associated with more reduced microbial proteins, informing about how the body's internal chemical environment changes in disease
(preprint: Dick, 2023).
Genomic adaptation to redox environments
Local and global chemical shaping of genomes. By combining taxonomic classifications with genome sequences, we can quantify genomic adaptation at the community level. Locally, carbon oxidation state (ZC) of community reference proteomes tracks oxygen concentration in hydrothermal systems, shale gas wells, and redox-stratified water bodies (Microbial Ecology, 2023 ). Globally, protein ZC is positively correlated with redox potential in aquatic, terrestrial, hydrothermal, and hyperalkaline environments, suggesting new ways in which genomic adaptation reflects physicochemical conditions (mSystems, 2023).
How can we get geochemical information from genomes? Distinct redox boundaries characterize evolutionary transitions in lineages of methanogens and ammonia-oxidizing Archaea. These boundaries are comparable to ranges of hydrogen activity found in modern ecosystems and could serve as proxies for past environments (Geobiology, 2023 ).
A role for water in cancer proteomes
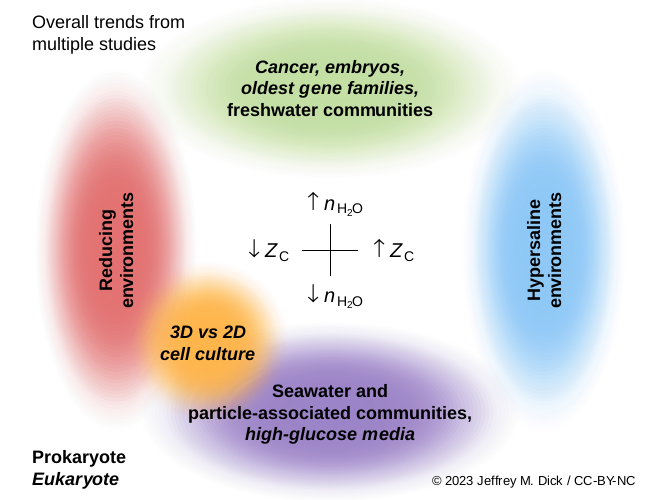
Stoichiometric hydration state (nH2O) is the amount of water in balanced chemical reactions that represent changes in protein expression levels. In an analysis of hundreds of proteomic datasets (available in the canprot package), cancer tissue generally exhibits higher proteomic nH2O than healthy tissue. In contrast, cells grown in high-glucose media or in 3D cell culture instead of monolayers are most often associated with relatively low proteomic nH2O (Computational and Systems Oncology, 2021). These are the first results to suggest a physicochemical link between protein expression patterns and cellular water content, which is elevated in tumors and proliferating cells.
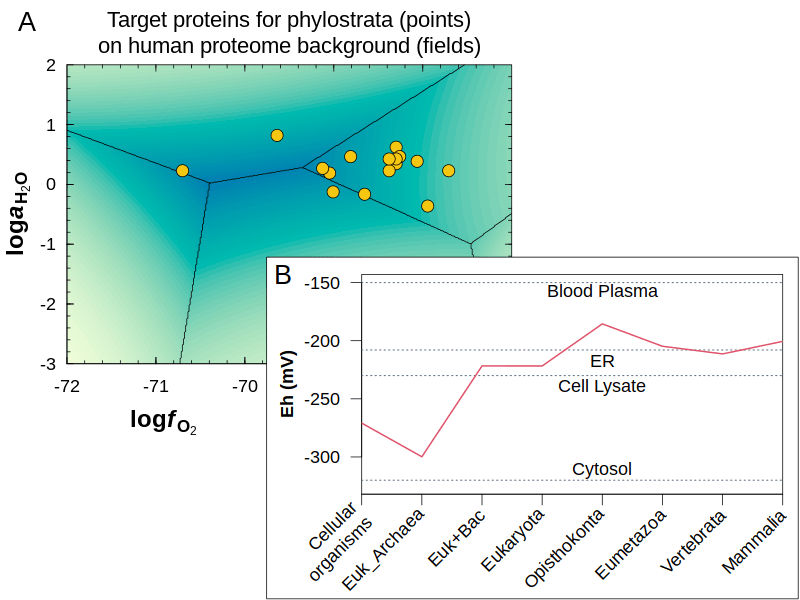
Connecting protein evolution to redox potential. A thermodynamic model was used to find chemical conditions that are most favorable for the formation of target proteins with different gene ages. Water activity and oxygen fugacity represent two dimensions of chemical variability (A), but the theoretical values are not directly comparable to measurements, so they were combined to calculate redox potential. This application of geochemical thermodynamics to sequence data provides a record of redox potential in deep time (B) that is consistent with a possible origin of the first cells in a reducing environment on Earth (Journal of Molecular Evolution, 2022 ). The data and code to make these and other plots from my papers are available in the JMDplots package.