Not what you're looking for? Try the first set of demos .
.
Hover over a description to show an image; click on the blue links to view the R code.
glycinate — Metal-glycinate complexes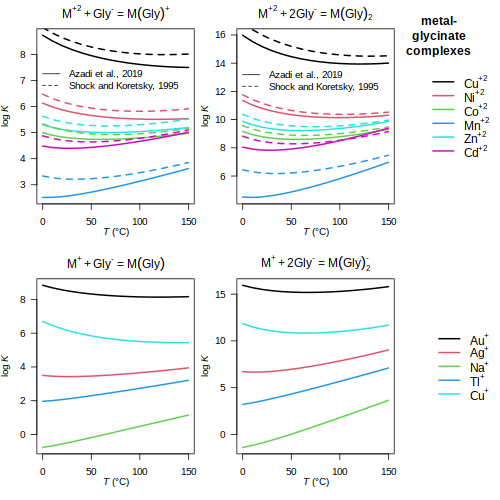
(
Azadi et al., 2019
;
Shock and Koretsky, 1995
)
copper — Complexation of copper with glycine species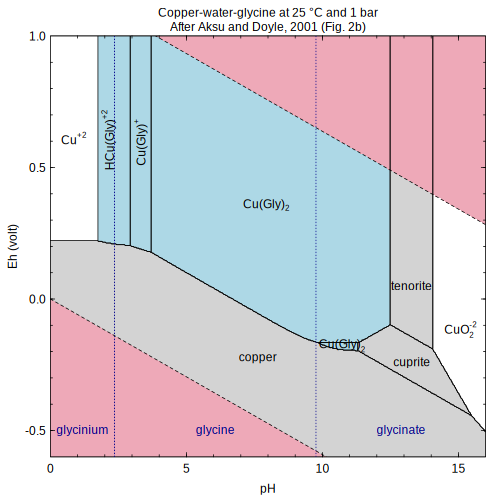 (Aksu and Doyle, 2001)
(Aksu and Doyle, 2001) affinity
affinity —
Amino acid synthesis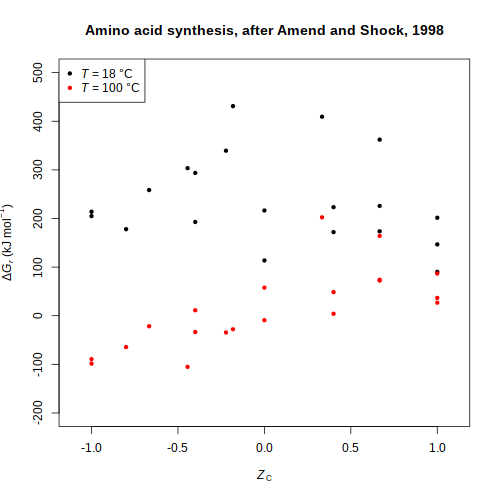
and
Aquifex metabolism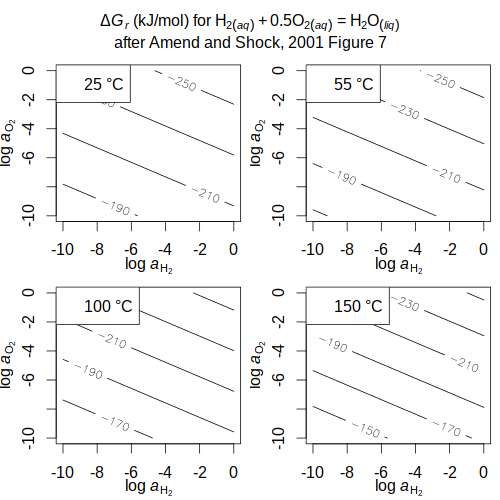 (Amend and Shock, 1998
(Amend and Shock, 1998
,
2001) buffer — Hydrogen fugacity for mineral buffers and aqueous species
buffer — Hydrogen fugacity for mineral buffers and aqueous species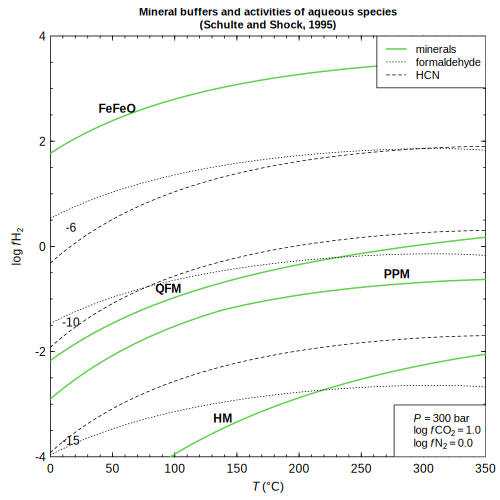 (Schulte and Shock, 1995)
(Schulte and Shock, 1995) ORP — Temperature dependence of oxidation-reduction potential for redox standards
ORP — Temperature dependence of oxidation-reduction potential for redox standards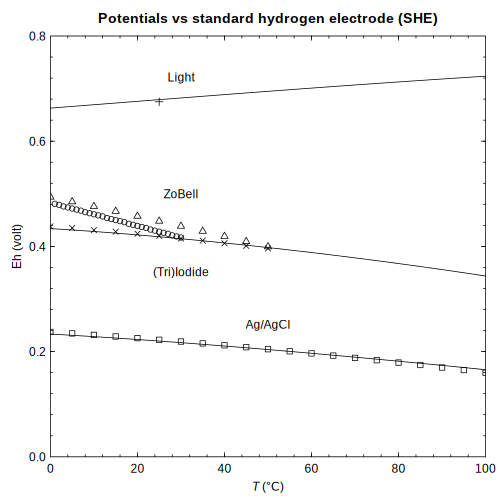 ionize — Contour plots of net charge and ionization properties of LYSC_CHICK
ionize — Contour plots of net charge and ionization properties of LYSC_CHICK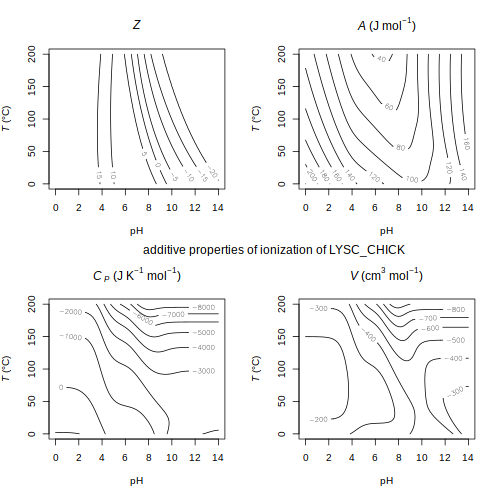 protbuff
protbuff — Hypothetical buffers of bacterial
thiol peroxidases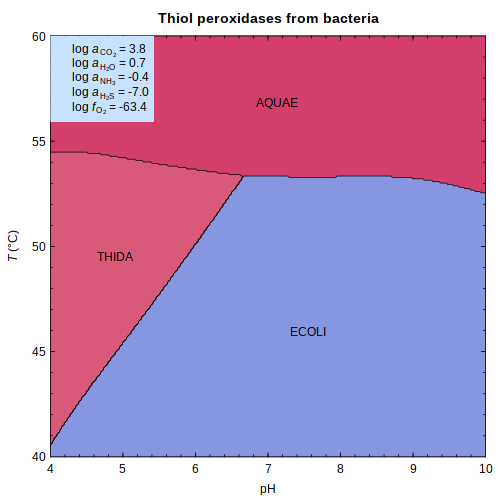
or
sigma factors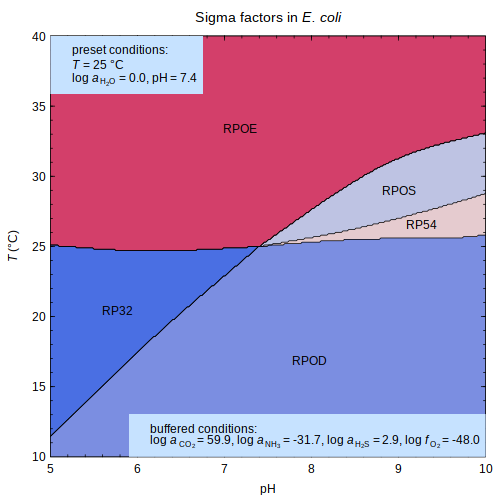
in
E. coli
carboxylase — Rank abundance distribution for RuBisCO and acetyl-CoA carboxylase (animated GIF)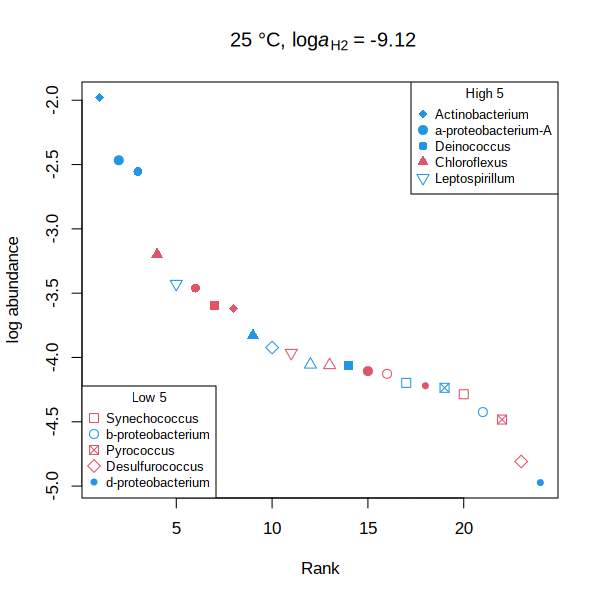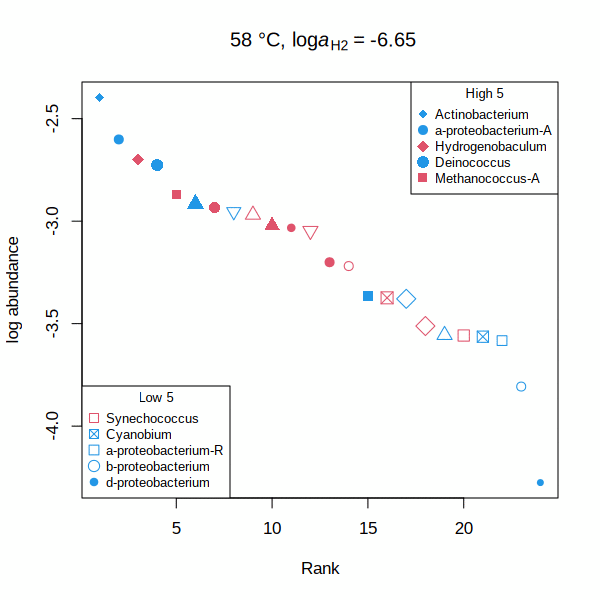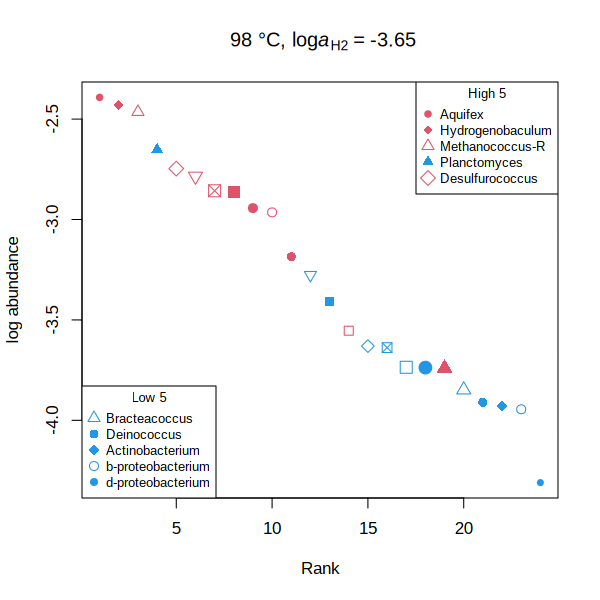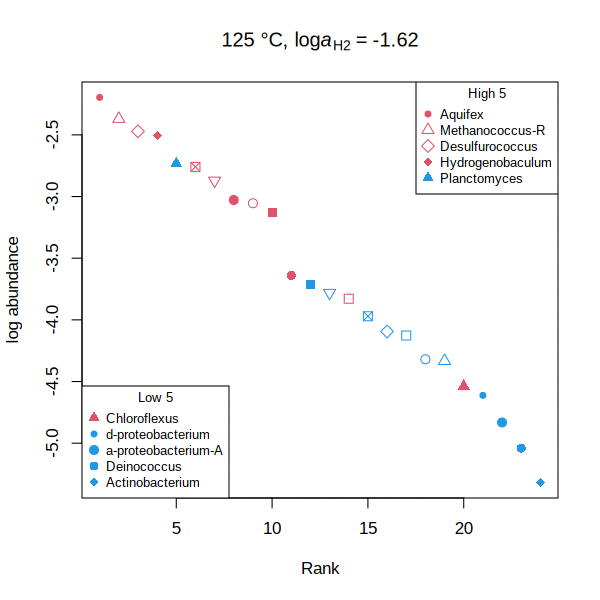 Shh — Affinities of transcription factors relative to Sonic hedgehog
Shh — Affinities of transcription factors relative to Sonic hedgehog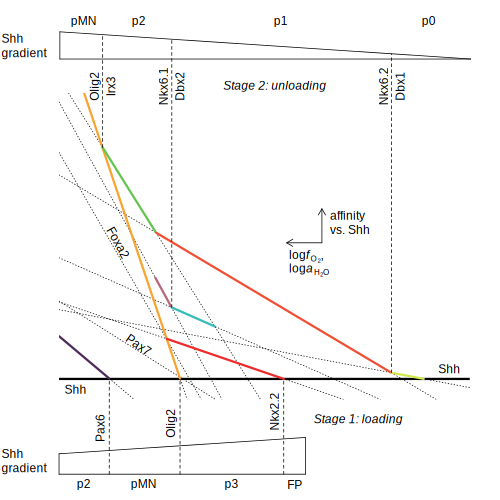 (Dick, 2015)
(Dick, 2015) dehydration — log K of dehydration reactions; SVG contains tooltips and links
dehydration — log K of dehydration reactions; SVG contains tooltips and links *
TCA — Standard Gibbs energies of steps of the citric acid cycle
*
TCA — Standard Gibbs energies of steps of the citric acid cycle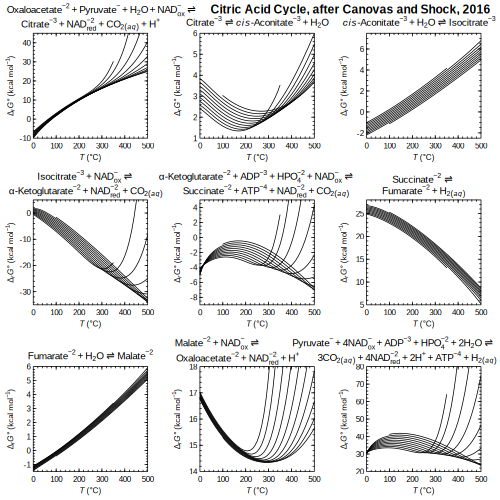 (Canovas and Shock, 2016)
(Canovas and Shock, 2016) adenine — HKF parameters regressed from heat capacity and volume
adenine — HKF parameters regressed from heat capacity and volume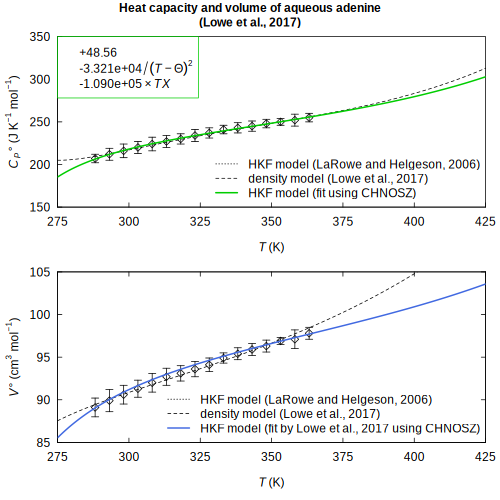 (Lowe et al., 2017)
(Lowe et al., 2017) comproportionation — Gibbs energy of sulfur comproportionation
comproportionation — Gibbs energy of sulfur comproportionation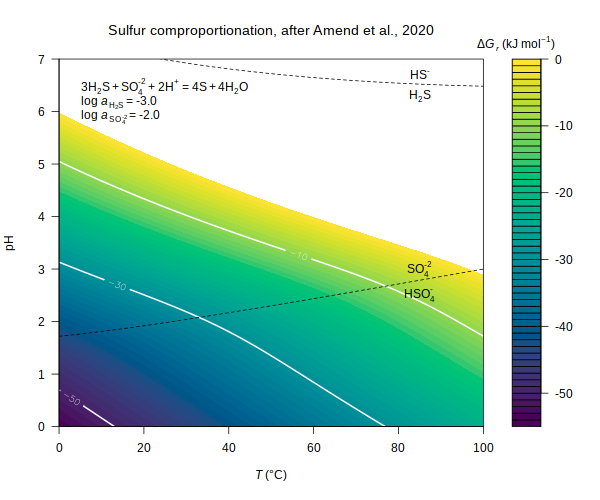 (Amend et al., 2020)
(Amend et al., 2020) E_coli — Gibbs energy of biomass synthesis in E. coli
E_coli — Gibbs energy of biomass synthesis in E. coli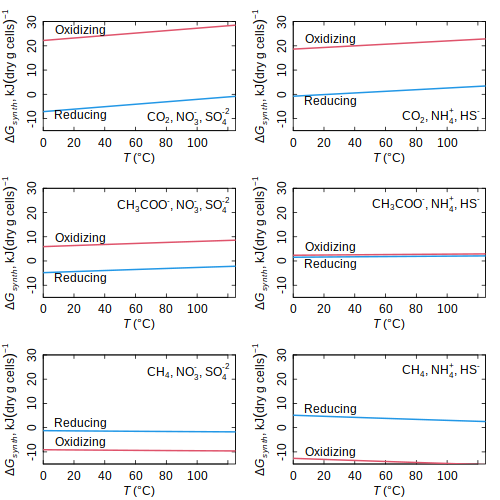 (LaRowe and Amend, 2016)
(LaRowe and Amend, 2016) rank.affinity — Affinity ranking for proteins
rank.affinity — Affinity ranking for proteins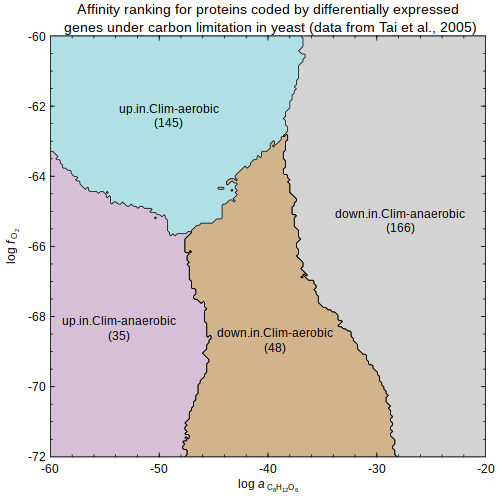
(gene expression data from
Tai et al., 2005
)
Last updated: 2025-06-25
 .
. .
. (Azadi et al., 2019
(Azadi et al., 2019 ; Shock and Koretsky, 1995
; Shock and Koretsky, 1995 )
) (Aksu and Doyle, 2001)
(Aksu and Doyle, 2001)
 and Aquifex metabolism
and Aquifex metabolism (Amend and Shock, 1998
(Amend and Shock, 1998 , 2001)
, 2001)
 (Schulte and Shock, 1995)
(Schulte and Shock, 1995)


 or sigma factors
or sigma factors in E. coli
in E. coli
 (Dick, 2015)
(Dick, 2015)
 *
* (Canovas and Shock, 2016)
(Canovas and Shock, 2016)
 (Lowe et al., 2017)
(Lowe et al., 2017)
 (Amend et al., 2020)
(Amend et al., 2020)
 (LaRowe and Amend, 2016)
(LaRowe and Amend, 2016)
 (gene expression data from Tai et al., 2005
(gene expression data from Tai et al., 2005 )
)